[Guidelines for Nomenclature 1]
Guidelines for Nomenclature of Genes, Genetic Markers, Alleles
1 Principles of Nomenclature
1.1 Key Features
The key component of nomenclature is the gene or locus name and symbol, which identifies a unit of inheritance. Other features, such as alleles, variants and mutations, are secondary to the gene name and become associated with it. Similarly, probes or assays used to detect a gene are not primary features and should not normally be used as names.
The primary purpose of a gene or locus name and symbol is to be a unique identifier so that information about the gene in publications, databases and other forms of communication can be unambiguously associated with the correct gene. These guidelines, therefore, are intended to aid the scientific community as a whole to use genetic information.
Other, secondary, functions of nomenclature for genes are to:
- identify the gene as a member of a family, which may give further information about the gene by reference to other family members
- identify the gene as the ortholog of a gene in another mammal (usually human)
1.2 Definitions
It is important that the user understands what is being named and the principles underlying these guidelines. Section 6 presents definitions that will aid the user in distinguishing, for example, genes, loci, markers, and alleles.
1.3 Stability of Nomenclature
On the whole gene names should be stable; that is, they should not be changed over time. However there are certain circumstances where a change is desirable:
- In cases where a gene has been known only as, and named for, a mutant phenotype: when the mutated gene is identified, then the mutant name becomes the mutant allele name of the identified gene (see Section 3.1.2).
- Where a gene becomes assigned to a gene family (of paralogs), and the nomenclature of the family is established. (see Section 2.6.2).
- Where orthologous gene(s) have been identified between mouse, rat, and human, and a common symbol is adopted for all three species.
1.4 Synonyms
A gene can have several synonyms, which are names or symbols that have been applied to the gene at various times. These synonyms may be associated with the gene in databases and publications, but the established gene name and symbol should always be used as the primary identifier.
1.5 Gene symbols, proteins, and chromosome designations in publications
1.5.1 Gene and allele symbols
Gene symbols are italicized when published, as are allele symbols. Section 2 below specifies naming rules for establishing correct symbols. Help is available for determining correct gene and allele symbol assignment (nomen@jax.org) and symbols can be reserved privately pre-publication.
To distinguish between mRNA, genomic DNA, and cDNA forms within a manuscript, write the relevant prefix in parentheses before the gene symbol, for example, (mRNA) Rbp1.
1.5.2 Protein symbols
Protein designations follow the same rules as gene symbols, with the following two distinctions:
- Protein symbols use all uppercase letters.
- Protein symbols are not italicized.
1.5.3 Chromosome designations
- Use uppercase "C" when referring to a specific mouse chromosome (e.g., Chromosome 15).
- When abbreviating the word Chromosome, do not use a period (".") after the abbreviation (e.g., Chromosome 15 should abbreviated as Chr 15 and not Chr.15).
2 Symbols and Names of Genes and Loci
The prime function of a gene name is to provide a unique identifier.
The Mouse Genome Database (MGD) serves as a central repository of gene names and symbols to avoid use of the same name for different genes or use of multiple names for the same gene (http://www.informatics.jax.org). The MGD Nomenclature Committee (nomen@jax.org) provides advice and assistance in assigning new names and symbols. A web tool for proposing a new mouse locus symbol is located at the MGD site.
For the rat, these functions are carried out by RGD (http://rgd.mcw.edu) assisted by the International Rat Genome and Nomenclature Committee (RGNC). A web tool for proposing a new rat locus symbol is located at the RGD site.
2.1 Laboratory Codes
A key feature of mouse and rat nomenclature is the Laboratory Registration Code or Laboratory code, which is a code of usually three to four letters (first letter uppercase, followed by all lowercase), that identifies a particular institute, laboratory, or investigator that produced, and may hold stocks of, for example, a DNA marker, a mouse or rat strain, or were the creator of a new mutation. Laboratory codes are also used in naming chromosomal aberrations, transgenes, and genetically engineered mutations. Because Laboratory codes are key to identifying original sources, they are not assigned to "projects," but rather to the actual producer/creator individual or site. Laboratory codes can be assigned through MGD or directly by the Institute for Laboratory Animal Research (ILAR) at http://dels-old.nas.edu/ilar_n/ilarhome/register_lc.php.
Examples:
J The Jackson Laboratory Mit Massachusetts Institute of Technology Leh Hans Lehrach Kyo Kyoto University Ztm Central Animal Laboratory Medical School Hannover
2.2 Identification of New Genes
Identification of new genes in general comes in two ways; identification of a novel protein or DNA sequence or identification of a novel phenotype or trait. In the case of sequences, care should be taken in interpretation of database searches to establish novelty (for example, to distinguish between a new member of a gene family and an allele or alternative transcript of an existing family member). Novel mutant phenotypes or traits should be named according to their primary characteristic, but once the gene responsible for the phenotypic variation is identified, this gives the primary name of the gene and the mutant name becomes the name of the allele (see Section 2.3).
2.3 Gene Symbols and Names
2.3.1 Gene Symbols
Genes are given short symbols as convenient abbreviations for speaking and writing about the genes.
A gene symbol should:
- be unique within the species and should not match a symbol in another species that is not a homolog.
- be short, normally 3-5 characters, and not more than 10 characters.
- use only Roman letters and Arabic numbers.
- begin with an uppercase letter (not a number), followed by all lowercase letters / numbers (see exception below).
- not include tissue specificity or molecular weight designations.
- include punctuation only in specific special cases (see below).
- ideally have the same initial letter as the initial letter of its gene name to aid in indexing. However, letter order in a gene symbol need not follow word order in the name.
Examples:
Plaur urokinase plasminogen activator receptor Sta autosomal striping
- be italicized in published articles. Because they may be difficult to read, depending on the browser, gene symbols are frequently not italicized when posted to a web page.
- use a common stem or root symbol when belonging to a gene family. Family member numbers or subunit designations should be placed at the end of the gene symbol.
Examples:
Glra1 glycine receptor, alpha 1 subunit Glra2 glycine receptor, alpha 2 subunit Glra3 glycine receptor, alpha 3 subunit
- use the same symbol whenever possible for orthologs among human, mouse and rat.
Exceptions to the rule of uppercase first letter and lowercase remaining letters in a gene or locus symbol:
- If the gene (locus) is only identified by a recessive mutant phenotype, then the symbol should begin with a lowercase letter. Once the mutant gene product is identified, the gene product is given a name and symbol and the original phenotype-based symbol and name becomes the allele symbol and name. The recessive nature of the allele is still conveyed by an initial lowercase letter.
- Within a gene symbol, Laboratory codes have an initial uppercase letter.
- When describing cross-hybridizing DNA segments, H (human) or other species code is uppercase, for example D2H11S14.
- When no information is available, other than the sequence itself, use the sequence identifier from the Mammalian Gene Collection, RIKEN, or GenBank (e.g., AF171077, 0610008A10Rik). If multiple sequence sources are available for the novel gene, preference is given first to a BC clone id (from Mammalian Gene Collection) followed by a RIKEN clone id, then the GenBank id.
Use of hyphens within the symbol should be kept to a minimum. Situations where hyphens may be used include:
- to separate related sequence and pseudogene symbols from the root
Examples:
Hk1-rs1 hexokinase-1 related sequence 1 Hba-ps3 hemoglobin alpha pseudogene 3
- within superscripted allele symbols (Section 3.1.2).
Example:
Kit W-v Kit oncogene
allele name: viable dominant spotting
2.3.2 Gene Names
Names of genes should be brief, and convey accurate information about the gene. The name should not convey detailed information about the gene or assay used; this can be associated with the gene in publications or databases. While the gene name should ideally be informative as to the function or nature of the gene, care should be taken to avoid putting inaccurate information in the name. For example, a "liver-specific protein" may be shown by subsequent studies to be expressed elsewhere.
A gene name should:
- be specific and brief, conveying the character or function of the gene.
- begin with a lowercase letter, unless it is a person's name or is a typically capitalized word.
- use American spelling.
- not contain punctuation, except where necessary to separate the main part of the name from modifiers.
- include the name of the species from which the ortholog/homolog name was derived at the end of the name in parentheses only when that name is not in common usage.
- not include the word mouse (for a mouse gene name) or the word rat (for a rat gene name).
- follow the conventions of the established gene family if it is a recognizable member of that family by sequence comparison, structure (motifs/domains), and/or function.
- not contain potentially misleading information that may be experiment or assay specific, such as "kidney-specific" or "59 kDa."
Examples:
Blr1 Burkitt lymphoma receptor 1 Acly ATP citrate lyase
Examples:
Acp1 acid phosphatase 1, soluble Pigq phosphatidylinositol glycan, class Q
Examples:
Shh sonic hedgehog
[commonly used, does not include species name]Fjx1 four jointed box 1 (Drosophila)
[name includes species derivative]
2.4 Structural Genes, Splice Variants, and Promoters
Ultimately, the majority of gene names will be for structural genes that encode protein. The gene should as far as possible be given the same name as the protein, whenever the protein is identified. If the gene is recognizable by sequence comparison as a member of an established gene family, it should be named accordingly (see Section 2.6).
2.4.1 Alternative Transcripts
Alternative transcripts that originate from the same gene are not normally given different gene symbols and names. To refer to specific splice forms of a gene, the following format should be used (gene symbol, followed by underscore, followed by sequence accession ID):
Example:
Gene Mttp microsomal triglyceride transfer protein Splice variant Mttp_EU553486 microsomal triglyceride transfer protein splice variant defined by transcript sequence EU553486
Using the sequence accession ID provides an unambiguous and precise definition to the splice variant.
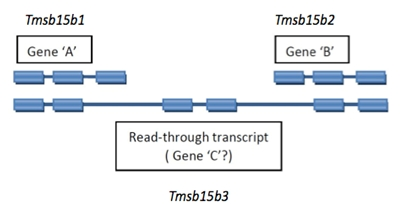
2.4.2 Read-through Transcripts
A read-through transcript is a multi-exon transcript that shares one of more exons with non-overlapping shorter transcripts that are considered to represent products of distinct loci. This is usually readily recognized as a distinct pattern, not to be confused with simple alternate splicing for a locus.
Read-through transcript genes should be named with a unique symbol and name. An example is diagrammed below.
2.4.3 Antisense and Opposite Strand Genes
Transcripts from the opposite strand that overlap another gene, or a transcript that is derived principally from the introns of another gene, or one that uses an alternative reading frame to another gene (and does not use the existing frame to a significant extent) should be given a different name.
A gene of unknown function, encoded at the same genomic locus (with overlapping exons) as another gene should have its own symbol. If the new gene regulates the first gene, it may be assigned the symbol of the first gene with the suffix “as” for antisense. The gene symbol should not be written backwards.
Example:
Igf2as insulin-like growth factor 2, antisense
Genes of unknown function on the opposite strand, which have no proven regulatory function, should be assigned the symbol of the known gene with the suffix “os” for opposite strand.
Example:
Dnm3os dynamin3, opposite strand
2.4.4 Genes with Homologs in Other Species
To aid interspecific comparison of genetic and other information, a gene that is identifiable as a homolog of an already named gene in another species can be named as "-like" "-homolog" or "-related." (Note: this is not the same as "related sequence" which applies to related sequences within mouse or within rat.) The gene name or symbol should not include the name mouse or the abbreviation "M" for mouse or the name rat or the abbreviation "R" for rat. Where possible, genes that are recognizable orthologs of already-named human genes should be given the same name and symbol as the human gene.
2.5 Phenotype Names and Symbols
Genes named for phenotypes should aim to convey the phenotype briefly and accurately in a few words. It is accepted that the name may not cover all aspects of the phenotype; what is needed is a succinct, memorable and, most importantly, unique, name. Bear in mind that identification of a variant or mutant phenotype is recognition of an allelic form of an as-yet unidentified gene that may already have or will be given a name.
2.5.1 Lethal Phenotypes
Genes identified solely by a recessive lethal phenotype with no heterozygous effect are named for the chromosomal assignment, a serial number and the name of the laboratory of origin (from the Laboratory code).
Examples:
l5H1 First lethal on Chromosome 5 at Harwell l4Rn2 Second lethal on Chromosome 4 from laboratory of Gene Rinchik
2.6 Gene Families
Genes that appear to be members of a family should be named as family members. Evidence of gene families comes in a variety of forms, e.g., from a probe detecting multiple bands on a Southern blot, but is principally based on sequence comparisons.
2.6.1 Families Identified by Hybridization
Historically, many gene families have been identified as fragments detected by hybridization to the same probe but which map to different loci. These family members may be functional genes or pseudogenes. The loci can be named "related sequence" of the founder gene with a serial number (symbol -rs1, -rs2, and so on).
Example:
mouse ornithine decarboxylase-related sequences 1 to 21. Odc-rs1 to Odc-rs21
If the founder or functional gene can not be identified, initially all the fragments are named "related sequence" until it is identified; then that particular "-rs" is dropped, without renumbering. If there is evidence that any loci are pseudogenes, they should be named as such and given serial numbers as in Section 2.6.2.
Once sequence evidence is accumulated on functional family members (which may or may not have been previously identified as members) a systematic naming scheme should be applied to the family as in Section 2.6.2.
2.6.2 Families Identified by Sequence Comparison
Sequencing can identify genes that are clearly members of a family (paralogs). Where possible, members of the family should be named and symbolized using the same stem followed by a serial number. The same family members in different mammalian species (orthologs) should, wherever possible, be given the same name and symbol. Pseudogenes should be suffixed by -ps and a serial number if there are multiple pseudogenes. Note that the numbering of pseudogenes among species is independent and no relationship should be implied among mouse, rat, or human pseudogenes based on their serial numbering.
Examples:
In mouse, phosphoglycerate kinase 1, pseudogenes 1 to 7, Pgk1-ps1 to Pgk1-ps7 In rat, calmodulin pseudogene 1, Calm-ps1
Numerous gene families have been recognized and given systematic nomenclature. Information on these families can be found at family-specific web sites, some of which are linked from MGD and RGD or RatMap. Names and symbols of new members of these families should follow the rules of the particular family and ideally be assigned in consultation with the curator of that family. Nomenclature schemes and curation of new families benefit from examination of existing models.
2.7 ESTs
Expressed Sequence Tags (ESTs) differ from other expressed sequences in that they are short, single pass sequences that are often convenient for PCR amplification from genomic DNA. ESTs that clearly derive from a known gene should be considered simply as an assay (marker) for that known gene. When anonymous ESTs are mapped onto genetic or physical maps, their designations should be symbolized using their sequence database accession number.
2.8 Anonymous DNA Segments
Only anonymous DNA segments that are mapped should be given systematic names and symbols.
2.8.1 Mapped DNA Segments
Anonymous DNA segments are named and symbolized according to the laboratory identifying or mapping the segment as "DNA segment, chromosome N, Lab Name" and a serial number, where N is the chromosomal assignment (1-19, X, Y in the mouse and 1-20, X, Y in the rat) and is symbolized as DNLabcode#.
Examples:
D8Mit17 the 17th locus mapped to mouse Chromosome 8 by M.I.T. D1Arb27 the 27th locus mapped to rat Chromosome 1 at the Arthritis and Rheumatism Branch, NIAMS.
The same convention is applied to DNA segments that are variant loci within known genes.
Examples:
D4Mit17 an SSLP within the mouse Orm1 gene D20Wox37 an SSLP within the rat Tnf gene
Mouse or rat DNA segments that are detected by cross-hybridization to human segments are given the human name with "chromosome N, cross-hybridizing to human DNA segment" inserted between DNA segment and the human segment code (see symbols). The same applies for rat DNA segments detected by cross-hybridization to mouse segments (or vice versa).
Examples:
D16H21S56 Mouse DNA segment on Chr 16 that cross-hybridizes with a DNA segment D21S56 from human Chr 21. D1M7Mit236 Rat DNA segment on Chr 1 that cross-hybridizes with a DNA segment D7Mit236 from mouse Chr 7
2.8.2 STSs Used in Physical Mapping
When physical maps are assembled (YAC or BAC contigs, for example) many markers may be placed on the map in the form of Sequence Tagged Sites (STSs). These might be clone end-fragments, inter-repeat sequence PCR products, or random sequences from within clones. These markers serve to validate the contigs and appear on the maps, but their further utility may be limited. It is not necessary to give them names or symbols other than those assigned by the laboratory that produced and used them. If the STSs are used more widely, they should be assigned anonymous DNA segment names ("D-numbers").
2.9 Gene Trap Loci
Gene trap experiments in embryonic stem (ES) cells produce cell lines in which integration into a putative gene is selected by virtue of its expression in ES cells. The trapped gene is usually (though not necessarily) mutated by the integration. The site of integration can be characterized by a number of means, including cloning or extension of cDNA products. The loci of integration of a series of gene trap lines, once characterized as potentially unique, can be named and symbolized as members of a series, using the prefix Gt (for gene trap), followed by a vector designation in parentheses, a serial number assigned by the laboratory characterizing the locus, and the laboratory ILAR code. For example, the 26th gene "trapped" by the ROSA vector in the laboratory of Phillip Soriano (Sor) is symbolized as:
- Gt(ROSA)26Sor
A gene trap designation becomes an allele of the gene into which it was inserted, once that gene is identified. For example, Gt(ST629)Byg is known to disrupt the netrin 1 (Ntn1) gene; thus the full allele designation for this gene trap mutation is Ntn1Gt(ST629)Byg. See also the examples of gene trap mutations in Section 3.5.2.
2.10 Quantitative Trait Loci, Resistance Genes, and Immune Response Genes
Differences between inbred strains and the phenotype of offspring of crosses between strains provide evidence for the existence of genes affecting disease resistance, immune response, and many other quantitative traits (quantitative trait loci, QTL). Evidence for QTL is generally obtained through extensive genetic crossing and analysis that may uncover many genetic elements contributing to a phenotypic trait. Generally, the number and effects of QTL can only be deduced following experiments to map them. QTL should not be named until such mapping experiments have been performed.
2.10.1 Names and Symbols of QTL
Names and symbols for QTL should be brief and descriptive and reflect the trait or phenotype measured. Those QTL affecting the same trait should be given the same stem and serially numbered. The series is separate for mouse and rat and no homology should be implied by the serial numbers.
Some historically named QTL carry the name of the disease with which they are associated; these names are maintained; but newly identified QTL should be named for the measured trait and not a disease. The suffix "q" may be used optionally as the final letter preceding the serial number in QTL symbols.
Naming and symbolizing QTL follow the same conventions as for naming and symbolizing genes (Section 2.3). Specifically for a QTL, its name should include:
- a root name describing the measured trait
- the designation QTL (recommended)
- a serial number
Examples:
in mouse Cafq1 caffeine metabolism QTL 1 Cafq2 caffeine metabolism QTL 2 Cafq3 caffeine metabolism QTL 3 in rat Kidm1 kidney mass QTL 1 Kidm2 kidney mass QTL 2 Kidm3 kidney mass QTL 3
To obtain the next available serial number for a new QTL with an already established root name, e.g., the next in the series of "liver weight QTL" in mouse (Lwq#) or the next in series of "blood pressure QTL" for rat (Bp#), users should submit their QTL on the "proposing a new locus symbol" form at MGD (for mouse) or RGD (for rat). Note that examining the database content for a QTL is not sufficient, as a laboratory may have a QTL designation reserved and private, pending publication.
2.10.2 Defining uniqueness in QTL
Specific circumstances for naming independent QTL include:
- Independent experiments study the same trait and map that trait to the same chromosomal region
Because QTL are detected in the context of specific strain combinations in specific crosses and generally in different laboratories using different assays, each experimentally detecting QTL will be given a unique symbol/name even when the trait measured and region defined is superficially the same as that of an existing QTL.
Example: In mouse, Obq1 (obesity QTL 1) was identified and mapped to Chromosome 7 in a cross between strains 129/Sv and EL/Suz. Another obesity QTL was also mapped to Chromosome 7, but because it involved distinct strains (NZO and SM), it was given a different QTL designation, Obq15.
- A chromosomal region containing many measured "traits"
If multiple traits are measured in a single experiment and mapped to a single chromosomal region, there may or may not be evidence that different QTL are involved. If the traits are physiologically related, the QTL name should be broad enough to represent all the measured traits or the name should reflect the trait showing the highest LOD score/p-value. Conversely, if there is clear evidence that the traits are independent, each trait will constitute a unique QTL.
Examples: In mouse, Nidd1 (non-insulin-dependent diabetes mellitus 1) was associated with related measurements of plasma insulin, non-fasted blood glucose, and body weight and given a single QTL designation.
In rats, Uae5 (urinary albumin excretion QTL 5) and Cm16 (cardiac mass QTL 16) are QTLs derived from the same experiment that map to overlapping regions of Chromosome 1. Because the measured traits are independent, different QTL designations are assigned.
2.11 Chromosomal Regions
Separate documents detail guidelines for nomenclature of chromosomes (for mouse, Rules for Nomenclature of Chromosome Aberrations are online; for rat, see Levan, et al., 1995). However, certain cytological features of normal chromosomes (such as telomeres, centromeres, and nucleolar organizers) and abnormal chromosomes (such as homogeneously-staining regions and end-points of deletions, inversions, and translocations) are genetic loci that are given names and symbols.
2.11.1 Telomeres
The functional telomere should be denoted by the symbol Tel. A DNA segment that includes the telomere repeat sequence (TTAGGG)n and which maps to a telomeric location is symbolized in four parts:
- Tel (for telomere)
- The number of the chromosome
- p or q (for the short or long arm, respectively)
- A serial number, if more than one segment is assigned to the telomere
For example, Tel4q1 telomeric sequence, Chr4, q arm 1
2.11.2 Centromeres and Pericentric Heterochromatin
The functional centromere should be denoted by the symbol Cen. Until the molecular nature of a functional mammalian centromere is defined, DNA segments that map to the centromere should be given anonymous DNA segment symbols as in Section 2.8.1.
Pericentric heterochromatin, that is cytologically visible, is given the symbol Hc#, in which # is the chromosome on which it is located.
- For example, Hc14 is the pericentric heterochromatin on Chromosome 14.
Variation in heterochromatin band size can be denoted by superscripts to the symbol.
- For example, Hc14n is normal heterochromatin; superscripts l and s would be used to denote long and short heterochromatin, respectively.
2.11.3 Nucleolus Organizers
The nucleolus organizer is a cytological structure that contains the ribosomal RNA genes. These genes are given the symbols Rnr and the number of the chromosome on which they are located.
- For example, Rnr12 is the ribosomal RNA locus on Chromosome 12.
If different Rnr loci can be genetically identified on the same chromosome, they are given serial numbers in order of identification.
- For example, Rnr19-1, Rnr19-2.
2.11.4 Homogeneously Staining Regions
Homogeneously staining regions (HSRs) are amplified internal subchromosomal bands that are identified cytologically by their Giemsa staining. A DNA segment that maps within an HSR is given a conventional DNA segment symbol, when its locus is on a normal (unamplified) chromosome. When expanded into an HSR its symbol follows the guidelines for insertions, thus becoming, for example, Is(HSR;1)1Lub.
2.11.5 Chromosomal Rearrangements
Symbols for chromosomal deletions, inversions, and translocations are given in the chromosomal nomenclature section. The end points of each of these rearrangements, however, define a locus. Where there is only a single locus on a chromosome, the chromosome anomaly symbol serves to define it. However, where an anomaly gives two loci on a single chromosome they can bedistinguished by the letters p and d for proximal and distal.
- For example, In(1)1Rk-p, In(1)1Rk-d are the proximal and distal end points of the chromosomal inversion In(1)1Rk in mouse.
2.12 Genes Residing on the Mitochondria
The mitochondria carry essential genes, among them many transfer RNA (tRNA) genes. Genes residing on the mitochondria have a prefix mt- (lowercase mt followed by a hyphen). For transfer RNAs, the symbols consist of three parts, mt-, T (for tRNA), and a single lowercase letter for the amino acid. The chromosomal designation for mitochondrial genes is Chr MT.
Examples:
mt-Tc tRNA, cysteine, mitochondrial
(a tRNA gene residing on the mitochondria)mt-Atp6 ATP synthase 6, mitochondrial
(a non-tRNA gene residing on the mitochondria)
2.13 RNA Genes Encoded in the Nucleus
There are hundreds of loci encoding transfer RNAs (tRNA) and ribosomal RNAs (rRNA), and many are encoded in the nucleus. The following method symbolizes these nuclear-encoded RNA genes:Naming nuclear encoded transfer-RNAs
Symbols for nuclear encoded transfer-RNAs consist of four parts:
n- lowercase n followed by a hyphen to indicate nuclear encoding T uppercase T to indicate transfer-RNA aa the single letter abbreviation for the amino acid # serial number for this transfer-RNA Example: n-Ta12 nuclear encoded tRNA alanine 12 (anticodon AGC)
Naming nuclear encoded ribosomal-RNAs
Symbols for nuclear encoded ribosomal-RNAs consist of four parts:
n- lowercase n followed by a hyphen to indicate nuclear encoding R uppercase R to indicate ribosomal-RNA subunit the subunit designation # serial number for this ribosomal-RNA Example: n-R5s104 nuclear encoded rRNA 5S 104
2.14 microRNAs and microRNA clusters
MicroRNAs (miRNAs) are abundant, short RNA molecules that are post-transcriptional regulators that bind to complementary sequences on target mRNA transcripts, usually resulting in translational repression or target degradation and gene silencing.
Naming microRNAs Symbols for microRNAs consist of the root symbol Mir followed by the numbering scheme tracked in the miRBase database (www.mirbase.org), a database tracking microRNAs reported for all species.
For example, mouse Mir143 (microRNA 143) is represented as mmu-mir-143 in miRBase, with the mmu signifying mouse.
Naming microRNA clusters A microRNA cluster consists of several microRNAs in immediate genome proximity. These may be given symbols and names to refer unambiguously to the entire cluster.
For a microRNA cluster, the name will consist of the root symbol Mirc (for microRNA cluster) followed by a serial number (1, 2, 3…) for the cluster. MGI (for mouse) or RGD (for rat) should be consulted for the next available cluster number when a new cluster is defined. The list of microRNAs included in each cluster will be recorded in relevant database records for the genes, knockouts, and strains.
(Note that this differs from the definition of miRBase, which simply refers to clustered miRNAs as those less than 10kb from the miRNA of interest. Thus, in miRBase clusters defined based on one miRNA may or may not overlap clusters based on another miRNA.
2.15 Enhancers, Promoters, and Regulatory Regions
Enhancers, promoters, and regulatory regions can influence multiple genes. In addition, they can be localized far away from the gene(s) that they affect. Thus, it is misleading to name them based on the gene for which regulation was first recognized.
Enhancers, promoters, and regulatory regions are to be symbolized as:
Rr# regulatory region # where # indicates the next number in the series.